At Buzzfeed we love our graphs. Since our posts get pretty popular we have some big ones to deal with. Even so; we want to know a lot of statistics about them right up to the second.
Our language of choice is Python. Most of the time when we work with graphs we use nice packages like igraph or networkx. These allow us to do complex ad-hoc reports and analysis. For real-time applications and APIs, however, loading the graph into memory on-demand is totally prohibitive, the data is too big.
We could use a specialized database, but it's still slower to compute an arbitrarily complex query than to do primary key lookup. In this post we'll start by scaling those in-memory computations with a low-level Python implementation of my favorite interview question. We'll end with computations that are fully online.
Step 1: We start with a simple little tree

The Adjacency Matrix

The recursive version
Ok! We've done it. get_descendants takes a root node, "node" as input, as well as the adjacency matrix A (we're using a scipy.sparse.coo_matrix here).
I'm not going to explain recursion here. This is the most common answer I see when I pose someone this question. Admittedly, it's really nice looking!
But there is a problem.
Step 2: Water your little tree, so it grows.
The Queue Version
It's pretty simple. A queue (a double-ended queue here) allows us to avoid recursion. This makes the code more efficient and scalable.
Step 3: Plenty of sunlight!
The Buffered Queue Version
The beauty! The simplicity! The queue of unexplored nodes can still grow kinda fast, but now we're not storing the whole subtree in memory before we pass it on to the calling context.
The Graph Approach
Ok, that was pretty neat. We've got a pretty efficient search going. But the adjacency matrix isn't the only approach to this problem.
Let's say we define a simple data structure called a "Node". It has a name, and it can have children. Since our graph is directed we're presuming we can't traverse "up", only "down" to descendants. Hence no parents.
If we use consecutive node names, then the graph becomes a really simple list of nodes. We can jump all around by accessing that list by the index corresponding to a node's name.
This procedure gives us the graph representation of the adjacency matrix.
I have to address the elephant in the room at this point. Guido's essay on graphs.
That structure is great. It's simple, readable, and efficient. The benefit of it over the version above is that it allows an arbitrary node name. We don't need that in our case, so we're sticking with lists which are slightly more light weight.
A familiar friend
The function above probably looks VERY familiar. It's exactly the same relatively scalable method we used above, except the method that accesses child nodes has changed. In this case we've gone from two dot operators to one. That change is pretty negligible, in case you're wondering.
How do our two approaches compare?

Taking it online!
Recently my brother and I did a talk at about.com where we went over this material. You can find some working code examples on my github here. The code in this post is from there.
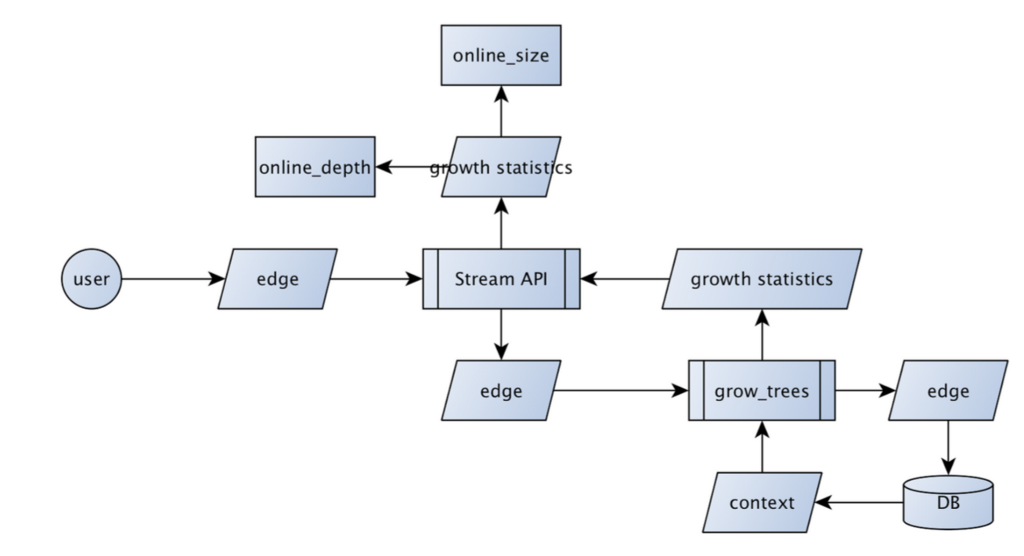
Parent and child are the names of the nodes included in the event. created_at was the time the event occurred, and generation represents the depth of the parent node in the tree.
The edge gets sent to a scalable messaging platform responsible for dispatching it to consumers. NSQ is among them, as well as Kafka, RabbitMQ, and even Redis.
Our consumer is a process called "grow_trees".
Disclaimer: for these message buses you normally write a function that handles a single message, acknowledges it's been processed, or rejects it. This process is daemonized, and managed by something like daemontools/svc or init.d.
Above you see we just wrap a stream that provides messages and implement a for loop to simulate messages being sent. This allows us to simulate a graph incoming from some real users in a dev environment.
The edges are unpacked in a fine-grained interface and instantiated to their coarse-grained counterparts before being passed around.
Building trees online is tricky when you can't guarantee messages are delivered in order. You have to define a criteria for when an incoming edge is rejected and when it will be incorporated into the existing tree/forest.
Running the online process has to be scalable. What happens when we have 10s of 1000s of requests coming in per second? We need global knowledge of state capable of scaling to handle this sort of load.
Here we're implementing redis to start out. Redis Cluster is a project gaining more traction for scaling redis. Redis itself is more than enough for most cases.
Notice both the operations are commutative. The order they're executed does not matter. This is a crucial requirement for computing graph statistics using this technique.
In our case; we use the criteria that the edge has to create a leaf, as well as a first-parent. E.g. If two parents are encountered for the same child the earliest edge is incorporated while the later edge is discarded (preserving the tree structure).
We use blinker.signal to fire an event received by each of our online methods when a node is added to the graph.
You're probably wondering now... if we're incorporating information from the global state (database) it's not technically online.
The online algorithms themselves compute the statistics they're responsible for, however, from statistics rather than the incoming edges by themselves. Each of those processes are, hence, online in isolation.
Computing the size of the graph from a database would require counting the number of nodes associated with some indexes. Similarly; computing the depth of the graph would require traversing and keeping track of the deepest generation somewhere. Using the online method we avoid any sort of aggregations.
